Gram-Schmidt Procedure for Signals#
Given a set of finite-energy signals \( \{s_m(t), m = 1, 2, ..., M\} \), the Gram-Schmidt orthogonalization procedure is used to construct an orthonormal set of waveforms.
Initial Step: Normalize the First Signal#
Starting with the first signal \( s_1(t) \), with energy \( E_1 \):
This creates the first orthonormal waveform \( \phi_1(t) \), which is \( s_1(t) \) normalized to unit energy.
The Next Step: Orthogonalize the Second Signal#
For the second signal \( s_2(t) \), compute its projection onto \( \phi_1(t) \):
Subtract this projection to create a signal orthogonal to \( \phi_1(t) \):
Normalize \( \gamma_2(t) \) to obtain the second orthonormal waveform:
where \( E_2 = \int_{-\infty}^{\infty} |\gamma_2(t)|^2 dt \) is the energy of \( \gamma_2(t) \).
A General Step: Orthogonalization of the \( k \)-th Signal#
For the \( k \)-th signal \( s_k(t) \), compute:
where:
Normalize \( \gamma_k(t) \) to obtain the \( k \)-th orthonormal waveform:
where \( E_k = \int_{-\infty}^{\infty} |\gamma_k(t)|^2 dt \).
Resultant Orthogonal Set of Waveforms
This process continues for all \( M \) signals \( \{s_m(t)\} \), resulting in \( N \leq M \) orthonormal waveforms. These waveforms span the signal space defined by the original set.
Example: Applying the Gram-Schmidt Procedure to Rectangular Signals#
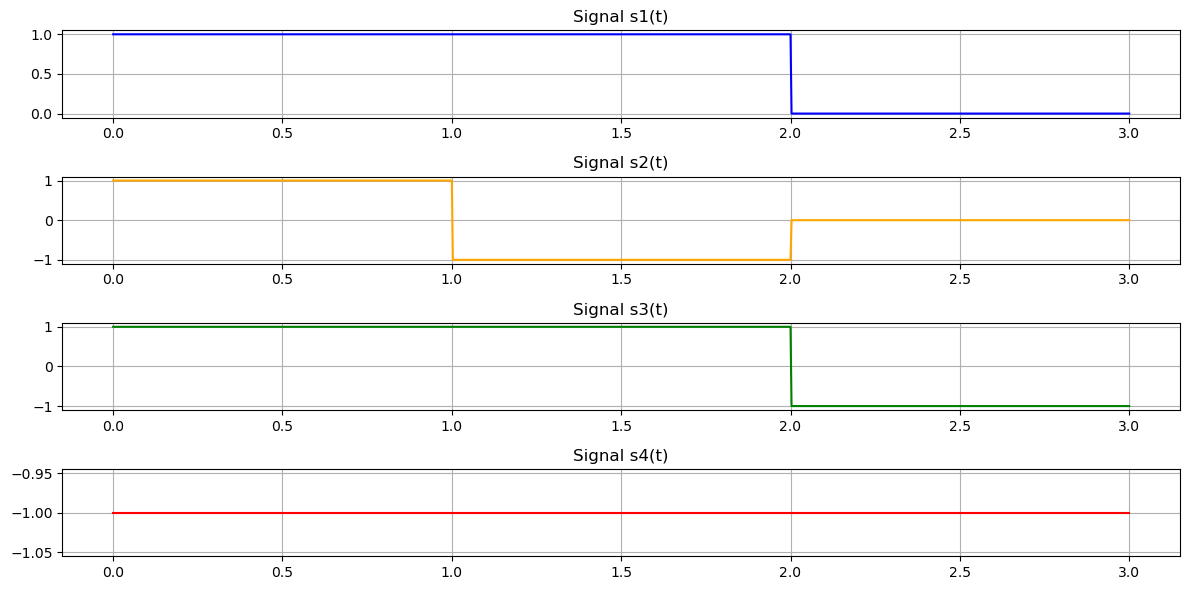
We apply the Gram-Schmidt process to four rectangular signals \( s_1(t), s_2(t), s_3(t), \) and \( s_4(t) \), where:
\( s_1(t) = \begin{cases} 1, & 0 \leq t \leq 2 \\ 0, & \text{otherwise} \end{cases} \)
\( s_2(t) = \begin{cases} 1, & 0 \leq t \leq 1 \\ -1, & 1 < t \leq 2 \\ 0, & \text{otherwise} \end{cases} \)
\( s_3(t) = \begin{cases} 1, & 0 \leq t \leq 2 \\ -1, & 2 < t \leq 3 \\ 0, & \text{otherwise} \end{cases} \)
\( s_4(t) = \begin{cases} -1, & 0 \leq t \leq 3 \\ 0, & \text{otherwise} \end{cases} \)
import numpy as np
import matplotlib.pyplot as plt
# Define time range
t = np.linspace(0, 3, 1000)
# Define the given signals
s1 = np.where((t >= 0) & (t <= 2), 1, 0)
s2 = np.where((t >= 0) & (t <= 1), 1, np.where((t > 1) & (t <= 2), -1, 0))
s3 = np.where((t >= 0) & (t <= 2), 1, np.where((t > 2) & (t <= 3), -1, 0))
s4 = np.where((t >= 0) & (t <= 3), -1, 0)
# Plot original signals
plt.figure(figsize=(12, 6))
plt.subplot(4, 1, 1)
plt.plot(t, s1, label="s1(t)", color="blue")
plt.title("Signal s1(t)")
plt.grid()
plt.subplot(4, 1, 2)
plt.plot(t, s2, label="s2(t)", color="orange")
plt.title("Signal s2(t)")
plt.grid()
plt.subplot(4, 1, 3)
plt.plot(t, s3, label="s3(t)", color="green")
plt.title("Signal s3(t)")
plt.grid()
plt.subplot(4, 1, 4)
plt.plot(t, s4, label="s4(t)", color="red")
plt.title("Signal s4(t)")
plt.grid()
plt.tight_layout()
plt.show()
Gram-Schmidt Procedure#
Step 1: Normalize \( s_1(t) \)
The energy of \( s_1(t) \) is:
The first orthonormal waveform is:
Step 2: Orthogonalize \( s_2(t) \)
The projection coefficient \( c_{21} \) is:
Since \( c_{21} = 0 \), \( s_2(t) \) and \( \phi_1(t) \) are orthogonal. The energy of \( s_2(t) \) is:
The normalized second orthonormal waveform is:
Step 3: Orthogonalize \( s_3(t) \)
The projection coefficients are:
The residual signal is:
The energy of \( \gamma_3(t) \) is:
The normalized third orthonormal waveform is:
Step 4: Orthogonalize \( s_4(t) \)
The projection coefficients are:
The residual signal is:
Since \( \gamma_4(t) = 0 \), \( s_4(t) \) is a linear combination of \( \phi_1(t) \) and \( \phi_3(t) \). Thus:
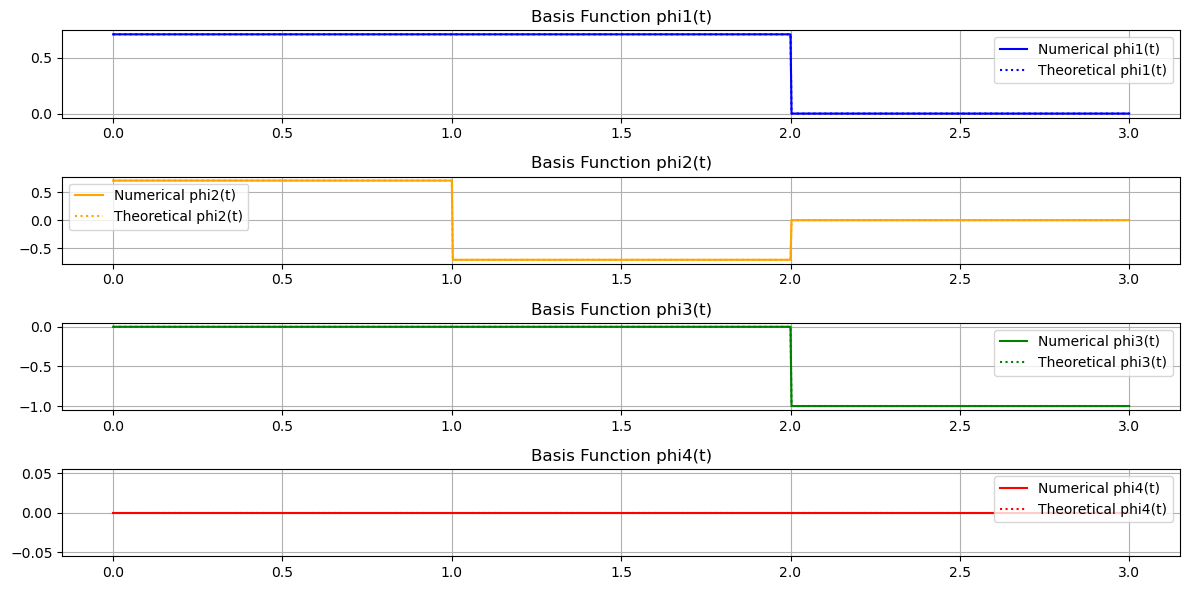
Final Results
\( \phi_1(t) = \frac{1}{\sqrt{2}} s_1(t) \)
\( \phi_2(t) = \frac{1}{\sqrt{2}} s_2(t) \)
\( \phi_3(t) = \gamma_3(t) = \begin{cases} -1 & \text{if } 2 \leq t < 3 \\ 0 & \text{otherwise} \end{cases} \)
\( \phi_4(t) = 0 \)
The final solution in the form of case functions:
\( \phi_1(t) \) (Normalized \( s_1(t) \)):
\( \phi_2(t) \) (Normalized \( s_2(t) \)):
\( \phi_3(t) \) (Orthogonalized \( s_3(t) \)):
\( \phi_4(t) \) (Resulting residual signal from \( s_4(t) \)):
# Step-by-step calculation of orthonormal waveforms
phi1 = s1 / np.sqrt(np.trapz(s1**2, t)) # Normalize s1(t)
c21 = np.trapz(s2 * phi1, t)
gamma2 = s2 - c21 * phi1
phi2 = gamma2 / np.sqrt(np.trapz(gamma2**2, t)) # Normalize gamma2
c31 = np.trapz(s3 * phi1, t)
c32 = np.trapz(s3 * phi2, t)
gamma3 = s3 - c31 * phi1 - c32 * phi2
phi3 = gamma3 / np.sqrt(np.trapz(gamma3**2, t)) # Normalize gamma3
c41 = np.trapz(s4 * phi1, t)
c42 = np.trapz(s4 * phi2, t)
c43 = np.trapz(s4 * phi3, t)
gamma4 = s4 - c41 * phi1 - c42 * phi2 - c43 * phi3
# Tolerance threshold for gamma4
tolerance = 1e-6
# Recalculate gamma4 with the threshold applied
if np.sqrt(np.trapz(gamma4**2, t)) < tolerance:
phi4 = np.zeros_like(t)
else:
phi4 = gamma4 / np.sqrt(np.trapz(gamma4**2, t))
# Define theoretical results
phi1_theoretical = np.where((t >= 0) & (t <= 2), 1/np.sqrt(2), 0)
phi2_theoretical = np.where((t >= 0) & (t <= 1), 1/np.sqrt(2), np.where((t > 1) & (t <= 2), -1/np.sqrt(2), 0))
phi3_theoretical = np.where((t > 2) & (t <= 3), -1, 0)
phi4_theoretical = np.zeros_like(t)
# Plot basis functions
plt.figure(figsize=(12, 6))
plt.subplot(4, 1, 1)
plt.plot(t, phi1, label="Numerical phi1(t)", color="blue")
plt.plot(t, phi1_theoretical, label="Theoretical phi1(t)", linestyle=":", color="blue")
plt.title("Basis Function phi1(t)")
plt.legend()
plt.grid()
plt.subplot(4, 1, 2)
plt.plot(t, phi2, label="Numerical phi2(t)", color="orange")
plt.plot(t, phi2_theoretical, label="Theoretical phi2(t)", linestyle=":", color="orange")
plt.title("Basis Function phi2(t)")
plt.legend()
plt.grid()
plt.subplot(4, 1, 3)
plt.plot(t, phi3, label="Numerical phi3(t)", color="green")
plt.plot(t, phi3_theoretical, label="Theoretical phi3(t)", linestyle=":", color="green")
plt.title("Basis Function phi3(t)")
plt.legend()
plt.grid()
plt.subplot(4, 1, 4)
plt.plot(t, phi4, label="Numerical phi4(t)", color="red")
plt.plot(t, phi4_theoretical, label="Theoretical phi4(t)", linestyle=":", color="red")
plt.title("Basis Function phi4(t)")
plt.legend()
plt.grid()
plt.tight_layout()
plt.show()
DISCUSSION
Why \( \phi_4(t) \) is Zero: Linear Dependence Explained
During the orthogonalization of \( s_4(t) \) against the orthonormal set \( \{\phi_1(t), \phi_2(t), \phi_3(t)\} \), \( \phi_4(t) \) was found to be zero (or effectively zero within numerical precision). This reflects the linear dependence among the signals.
Linear Dependence Overview:
A set of functions is linearly dependent if one function can be expressed as a linear combination of others.
If \( s_4(t) \) is a combination of \( s_1(t), s_2(t), \) and \( s_3(t) \), it does not contribute a new dimension to the signal space.
In the Gram-Schmidt Process:
The span of \( \{s_1(t), s_2(t), s_3(t)\} \) includes \( s_4(t) \), meaning \( s_4(t) \) is redundant.
Orthogonalizing \( s_4(t) \) yields \( \phi_4(t) = 0 \) because it lies within the span of the existing orthonormal functions.
Conclusion: Only \( \{\phi_1(t), \phi_2(t), \phi_3(t)\} \) form the orthonormal basis, as \( \phi_4(t) \) does not contribute additional information.
