Example 5.5#
This example serves to introduce the concept of signal space, which is used extensively to understand and interpret communications and radar-type detection problems from a geometrical point of view.
To start the understanding, consider two potential noise-free signals:
\[
u_0(t) = 1, \quad 0 \leq t \leq 1, \quad \text{hypothesis } H_0
\]
\[
u_1(t) = e^{j \beta}, \quad 0 \leq t \leq 1, \quad \text{hypothesis } H_1
\]
Thus, the two signals have equal energy:
\[
\mathcal{E}_0 = \mathcal{E}_1 = \frac{1}{2} \int_0^T |u_0(t)|^2 dt = \frac{1}{2} \int_0^T |u_1(t)|^2 dt = \frac{T}{2}
\]
and the correlation between them is:
\[
\rho = \frac{1}{T} \int_0^T e^{-j \beta} \, dt = e^{-j \beta}
\]
The error rate using these signals embedded in additive white Gaussian noise is
\[
P(E) = \frac{1}{2} \, \text{erfc} \left( \sqrt{\frac{\mathcal{E} (1 - \cos \beta)}{2 N_0}} \right)
\]
We can examine the performance with different signal-space representation of the two signals under different \( \beta \).
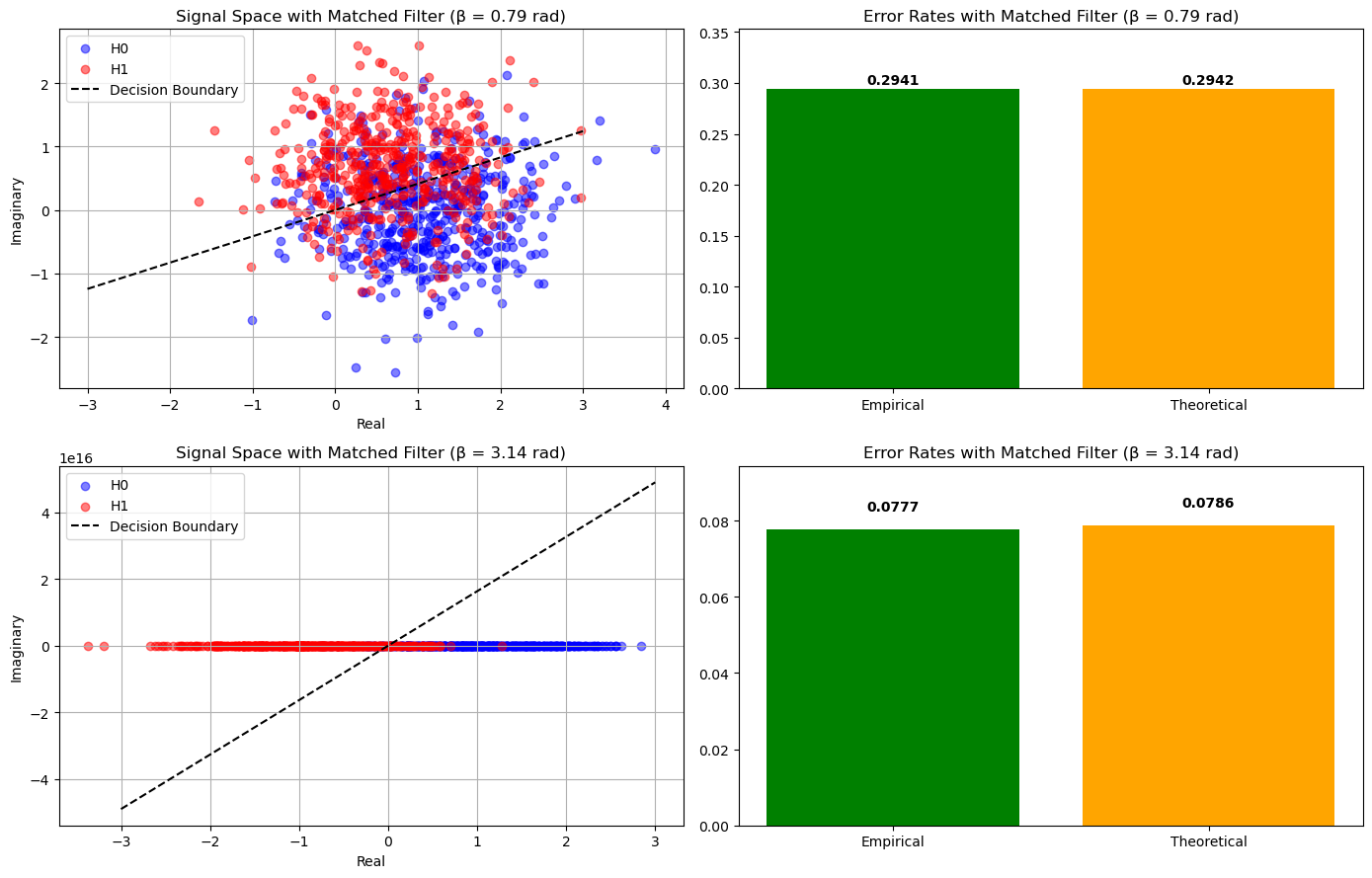
Simulation using Matched Filter#
import numpy as np
import matplotlib.pyplot as plt
from scipy.special import erfc
# Simulation Parameters
T = 1 # Duration
E = 0.5 # Energy
N0 = 1 # Noise power
num_trials = 100000 # Number of trials
beta_values = [np.pi / 4, np.pi ] # Beta values to simulate
# Function to generate noise
def generate_noise(num_samples):
# AWGN: real and imaginary parts are Gaussian with variance N0/2
noise_real = np.random.normal(0, np.sqrt(N0 / 2), num_samples)
noise_imag = np.random.normal(0, np.sqrt(N0 / 2), num_samples)
return noise_real + 1j * noise_imag
# Function to compute theoretical error rate
def theoretical_error(E, beta, N0):
# Must be 2E for COMPLEX BASEBAND SIGNAL
argument = np.sqrt((2*E * (1 - np.cos(beta))) / (2 * N0))
return 0.5 * erfc(argument)
# Function to perform simulation using matched filter for a given beta
def simulate_matched_filter(beta):
# Define signals
u0 = 1 # Hypothesis H0
u1 = np.exp(1j * beta) # Hypothesis H1
# Generate random hypotheses
# Assign H0 and H1 with equal probability
hypotheses = np.random.choice(['H0', 'H1'], size=num_trials)
transmitted_signals = np.where(hypotheses == 'H0', u0, u1)
# Add noise
noise = generate_noise(num_trials)
received_signals = transmitted_signals + noise
# Apply matched filters
# Matched filter for H0: h0(t) = u0*(t) = 1
# Matched filter for H1: h1(t) = u1*(t) = e^{-j*beta}
y0 = received_signals * 1 # Since h0(t) = 1
y1 = received_signals * np.exp(-1j * beta) # h1(t) = e^{-j*beta}
# Integrate over time T (since u0 and u1 are constant over [0, T])
# In discrete simulation, integration is equivalent to multiplication by T=1
# So, y0 and y1 are already the integrals
# Take real parts
y0_real = np.real(y0)
y1_real = np.real(y1)
# Decision rule: Decide H0 if Re(y0) > Re(y1), else H1
decisions = np.where(y0_real > y1_real, 'H0', 'H1')
# Compute error rate
errors = (decisions != hypotheses)
empirical_error = np.mean(errors)
# Compute theoretical error rate
theo_error = theoretical_error(E, beta, N0)
# Prepare data for signal space plot (only a subset for visualization)
subset = 1000 # Number of points to plot
received_subset = received_signals[:subset]
hypotheses_subset = hypotheses[:subset]
decisions_subset = decisions[:subset]
return empirical_error, theo_error, received_subset, hypotheses_subset, decisions_subset
# Plot settings
fig, axes = plt.subplots(2, 2, figsize=(14, 9))
for idx, beta in enumerate(beta_values):
empirical_error, theo_error, received_subset, hypotheses_subset, decisions_subset = simulate_matched_filter(beta)
print(f"Beta = {beta} radians")
print(f"Theoretical Error Rate: {theo_error:.4f}")
print(f"Empirical Error Rate: {empirical_error:.4f}\n")
# Plot signal space
ax1 = axes[idx, 0]
ax2 = axes[idx, 1]
# Plot received signals with different colors for H0 and H1
H0_indices = hypotheses_subset == 'H0'
H1_indices = hypotheses_subset == 'H1'
ax1.scatter(received_subset[H0_indices].real, received_subset[H0_indices].imag,
color='blue', alpha=0.5, label='H0')
ax1.scatter(received_subset[H1_indices].real, received_subset[H1_indices].imag,
color='red', alpha=0.5, label='H1')
# Plot decision boundary
# The decision boundary is defined by Re(y0) = Re(y1)
# Which implies Re(r) = Re(r e^{-j beta})
# To visualize, we can plot the line where Re(r) - Re(r e^{-j beta}) = 0
# Simplify: Re(r) - Re(r e^{-j beta}) = 0
# Let r = x + jy
# Re(r e^{-j beta}) = x cos(beta) + y sin(beta)
# So, x - (x cos(beta) + y sin(beta)) = 0 => x (1 - cos(beta)) - y sin(beta) = 0
# This is a straight line: y = (1 - cos(beta)) / sin(beta) * x
if beta != 0:
slope = (1 - np.cos(beta)) / np.sin(beta)
x_vals = np.linspace(-3, 3, 1000)
y_vals = slope * x_vals
ax1.plot(x_vals, y_vals, 'k--', label='Decision Boundary')
ax1.set_title(f'Signal Space with Matched Filter (β = {beta:.2f} rad)')
ax1.set_xlabel('Real')
ax1.set_ylabel('Imaginary')
ax1.legend()
ax1.grid(True)
# Plot theoretical vs empirical error
ax2.bar(['Empirical', 'Theoretical'], [empirical_error, theo_error],
color=['green', 'orange'])
ax2.set_ylim(0, max(empirical_error, theo_error)*1.2)
ax2.set_title(f'Error Rates with Matched Filter (β = {beta:.2f} rad)')
for i, v in enumerate([empirical_error, theo_error]):
ax2.text(i, v + 0.005, f"{v:.4f}", ha='center', fontweight='bold')
plt.tight_layout()
plt.show()
Beta = 0.7853981633974483 radians
Theoretical Error Rate: 0.2942
Empirical Error Rate: 0.2941
Beta = 3.141592653589793 radians
Theoretical Error Rate: 0.0786
Empirical Error Rate: 0.0777
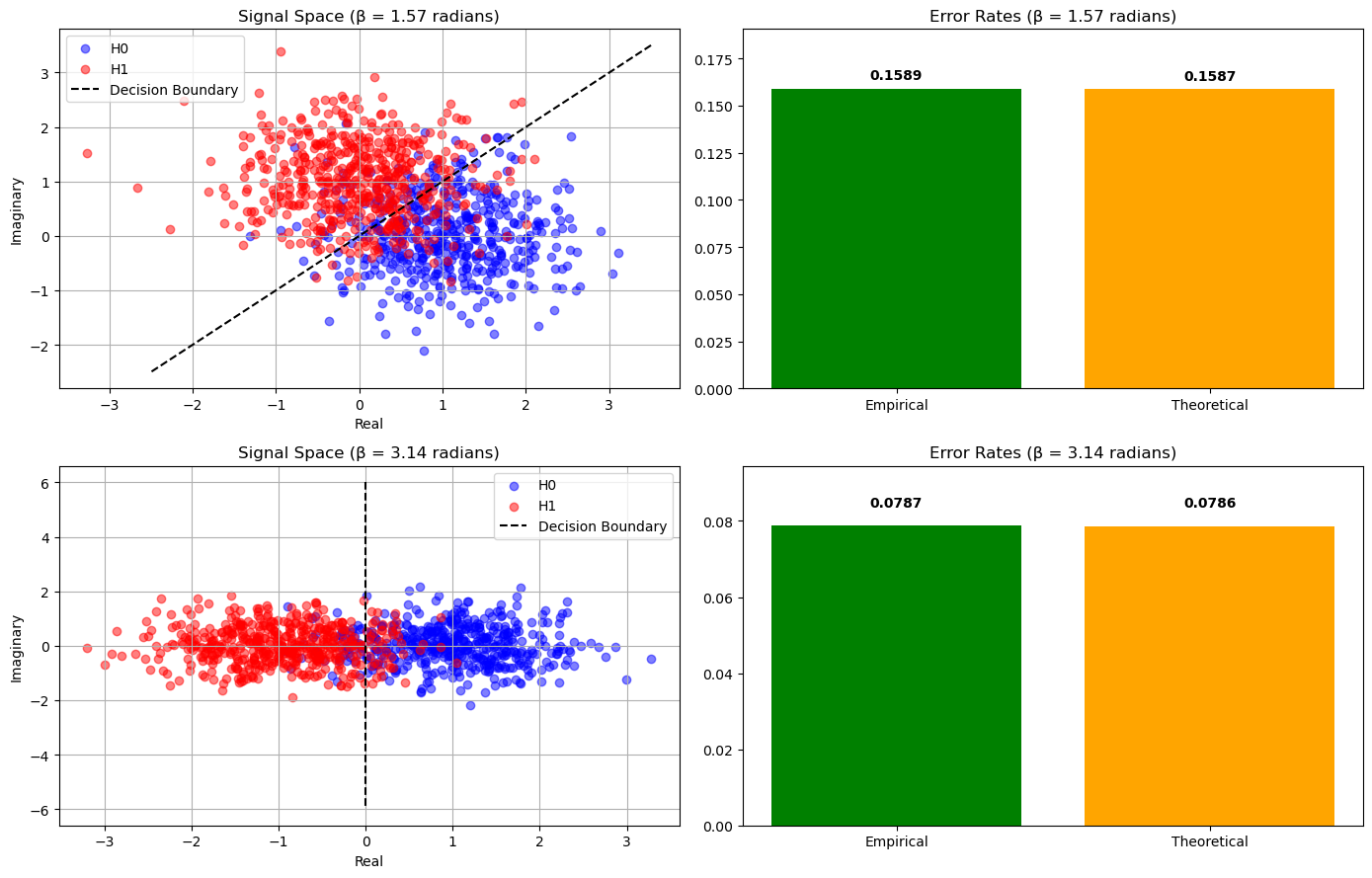
Simulation using Euclidean Distance#
import numpy as np
import matplotlib.pyplot as plt
from scipy.special import erfc
# Simulation Parameters
T = 1 # Duration
E = 0.5 # Energy
N0 = 1 # Noise power
num_trials = 1000000 # Number of trials
beta_values = [np.pi / 2, np.pi] # Beta values to simulate
# Function to generate noise
def generate_noise(num_samples):
# AWGN: real and imaginary parts are Gaussian with variance N0/2
noise_real = np.random.normal(0, np.sqrt(N0 / 2), num_samples)
noise_imag = np.random.normal(0, np.sqrt(N0 / 2), num_samples)
return noise_real + 1j * noise_imag
# Function to compute theoretical error rate
def theoretical_error(E, beta, N0):
# Must be 2E for COMPLEX BASEBAND SIGNAL
argument = np.sqrt((2*E * (1 - np.cos(beta))) / (2 * N0))
return 0.5 * erfc(argument)
# Function to perform simulation for a given beta
def simulate(beta):
# Define signals
u0 = 1 # Hypothesis H0
u1 = np.exp(1j * beta) # Hypothesis H1
# Generate random hypotheses
hypotheses = np.random.choice(['H0', 'H1'], size=num_trials)
transmitted_signals = np.where(hypotheses == 'H0', u0, u1)
# Add noise
noise = generate_noise(num_trials)
received_signals = transmitted_signals + noise
# Decision rule: Choose H0 if closer to u0, else H1
# Compute Euclidean distance in signal space
distance_H0 = np.abs(received_signals - u0)**2
distance_H1 = np.abs(received_signals - u1)**2
decisions = np.where(distance_H0 < distance_H1, 'H0', 'H1')
# Compute error rate
errors = (decisions != hypotheses)
empirical_error = np.mean(errors)
# Compute theoretical error rate
theo_error = theoretical_error(E, beta, N0)
# Prepare data for signal space plot (only a subset for visualization)
subset = 1000 # Number of points to plot
received_subset = received_signals[:subset]
hypotheses_subset = hypotheses[:subset]
decisions_subset = decisions[:subset]
return empirical_error, theo_error, received_subset, hypotheses_subset, decisions_subset
# Plot settings
fig, axes = plt.subplots(2, 2, figsize=(14, 9))
for idx, beta in enumerate(beta_values):
empirical_error, theo_error, received_subset, hypotheses_subset, decisions_subset = simulate(beta)
print(f"Beta = {beta} radians")
print(f"Theoretical Error Rate: {theo_error:.4f}")
print(f"Empirical Error Rate: {empirical_error:.4f}\n")
# Plot signal space
ax1 = axes[idx, 0]
ax2 = axes[idx, 1]
# Plot received signals with different colors for H0 and H1
H0_indices = hypotheses_subset == 'H0'
H1_indices = hypotheses_subset == 'H1'
ax1.scatter(received_subset[H0_indices].real, received_subset[H0_indices].imag,
color='blue', alpha=0.5, label='H0')
ax1.scatter(received_subset[H1_indices].real, received_subset[H1_indices].imag,
color='red', alpha=0.5, label='H1')
# Plot decision boundary
# The decision boundary is the perpendicular bisector between u0 and u1
# Compute midpoint
midpoint = (1 + np.exp(1j * beta)) / 2
# Compute the normal vector
normal = 1j * (np.exp(1j * beta) - 1)
# Generate a line for decision boundary
t = np.linspace(-3, 3, 1000)
boundary = midpoint + normal * t
ax1.plot(boundary.real, boundary.imag, 'k--', label='Decision Boundary')
ax1.set_title(f'Signal Space (β = {beta:.2f} radians)')
ax1.set_xlabel('Real')
ax1.set_ylabel('Imaginary')
ax1.legend()
ax1.grid(True)
# Plot theoretical vs empirical error
ax2.bar(['Empirical', 'Theoretical'], [empirical_error, theo_error],
color=['green', 'orange'])
ax2.set_ylim(0, max(empirical_error, theo_error)*1.2)
ax2.set_title(f'Error Rates (β = {beta:.2f} radians)')
for i, v in enumerate([empirical_error, theo_error]):
ax2.text(i, v + 0.005, f"{v:.4f}", ha='center', fontweight='bold')
plt.tight_layout()
plt.show()
Beta = 1.5707963267948966 radians
Theoretical Error Rate: 0.1587
Empirical Error Rate: 0.1589
Beta = 3.141592653589793 radians
Theoretical Error Rate: 0.0786
Empirical Error Rate: 0.0787